CQNR: Cluster Quality based Non-Reductional
over-sampling technique and EPP3D: Effector Protein
Predictor based on 3D structure of proteins
Rishika Sen, Somnath Tagore, Rajat K. De
CQNR - The phenomenon of the imbalanced dataset is common in classification. The existence of
this problem is visible in many real-life domains, especially for biological datasets. Data imbalance
problem can be tackled either by reducing the samples of the majority class or increasing
the number of samples in the minority class. In this paper, we have introduced Cluster Quality
based Non-Reductional over-sampling technique (CQNR). CQNR generates new samples
proportional to the distribution of samples of the minority classes, without eliminating any
sample of the minority class as noise.
EPP3D - Effector proteins of bacteria infect their hosts
by certain dedicated machinery, otherwise known as secretion systems. Therefore, we have
created a unique feature set consisting of eight features i.e., convex hull layer count, surface
atom composition, radius of gyration, packing density and compactness, derived from the 3D
structure of the experimentally verified effector proteins for its classification. For the classification
of pathogenic effector proteins, a significant improvement of approximately 5% has
been noticed, which has applied CQNR compared to the other oversampling methods. Based
on this feature set oversampled by CQNR, we have developed EPP3D, an effector protein
predictor based consensus of classifiers using majority voting.
Technical Details
CQNR and EEP3D are both python scripts that runs on Python 3.6 and above. It has been written using Win
Python in 64-bit version of Windows 8.1 operating system and a 32GB RAM. CQNR can read unbalanced dataset in csv format (comma separated).
The input unbalanced and the output balanced feature sets have rows as samples and columns as features.
The python packages needed for executing CQNR and EPP3D are
tkinter, random, pandas, numpy, csv, sklearn, keras, tensorflow,
collections, matplotlib, scipy, imblearn, decimal, warnings and re.
Downloads
Download the CQNR and EPP3D and its prerequisites from this link:
CQNR Package. Click the tab "clone or download".
On unzipping the package the following files/folders can be found:
- cqnr_sample - it has the datasets in csv format on which the performance of CQNR has been tested
- epp3d_sample_protein - it has the pdb files of proteins (effectors and non-effectors) on which
the performance of EPP3D has been tested
- CQNR.py - source code of CQNR in python
- EPP3D.py - source code of EPP3D in python
- all8features.csv - unbalanced eight feature set of the effector and non-effector protein
- training_epp3d.csv - balanced eight feature set of the effector and non-effector protein using which
EPP3D has to be trained
How to execute CQNR
Follow the steps to execute CQNR:
- Open WinPython-64bit-3.6.1.0Qt5 on Windows and open
the application IDLEX (Python Gui).
- From The GUI click "Open" which will open the file explorer.
- Find the file CQNR (if should be on the same folder as WinPython)
and click "Open" and "Run". On the IDE type "CQNR()" and click "Enter". A file
entry box will appear.
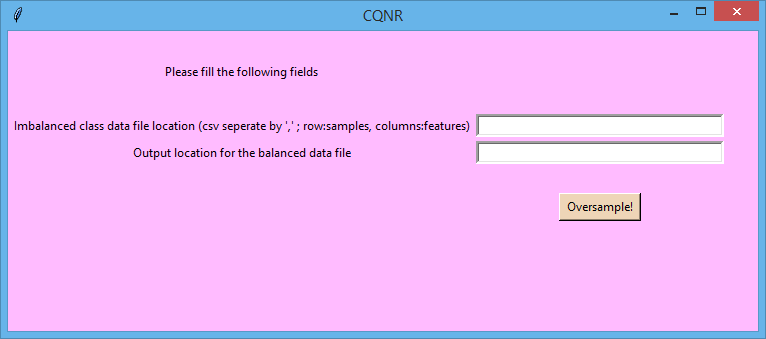
- For field "Imbalanced class data file location" give the path of the imbalanced dataset
(present in folder cqnr_sample) in CSV format.
- For field "Output location for the balanced data file" (in fasta format) give the path for
the balanced datset that will be produced by the algorithm.
- Fill the fields and click 'Oversample!'. Wait for the results.
How to execute EPP3D
Follow the steps to execute CQNR:
- Open WinPython-64bit-3.6.1.0Qt5 on Windows and open
the application IDLEX (Python Gui).
- From The GUI click "Open" which will open the file explorer.
- Find the file EPP3D.py.
and click "Open" and "Run". On the IDE type "EPP3D()" and click "Enter". A file
entry box will appear.
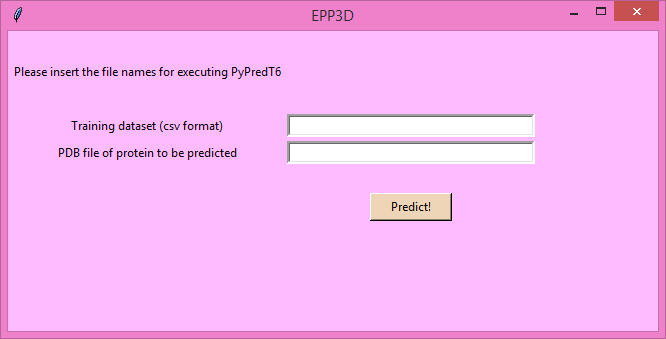
- For field "Training dataset" give the path for the training dataset (training_epp3d.csv) given in CSV format.
- For field "PDB file of protein to be predicted" (present in folder epp3d_sample_protein)
give the path for PDB file of the unknown protein that need to be classified.
- Fill the fields and click 'Predict!'. Wait for the results.


