PyPredT6: A Python based Prediction Tool for Identification of Type VI Effector
Proteins
Rishika Sen, Losiana Nayak, Rajat K. De
rishika21sen@gmail.com, losiana_t@isical.ac.in, rajat@isical.ac.in
Effector proteins of bacteria infect their hosts by certain dedicated machinery present in them,
otherwise known as Secretion System (SS). So far six such Secretion Systems (SS) have been identified
in gram-negative bacteria, i.e., T1SS -T6SS. T6 effector proteins of many gram-negative bacteria
have not yet been discovered. We have developed PyPredT6, a Python-based tool which provides
a convenient way to predict whether a protein is a T6 effector or not.
We have taken experimentally validated effector proteins and
designed a classification system based on a consensus of classifiers. We have used the predicted
result of Artificial Neural Network (ANN), Support Vector Machine (SVM), k Nearest Neighbors
(kNN), Naive Bayes (NB) and Random Forest (RF) to annotate an unknown protein.
Technical Details
PyPredT6 is a python script that runs on Python 3.6 and above. It has been written using Win
Python in 64-bit Windows 8.1 operating system and a 32GB RAM. PyPredT6 can read nucleotide and amino acid sequences from text
files in fasta format. The secondary structures from the amino acid sequences of the proteins
have been extracted from PaleAle 4.0 tool online. The python packages needed for executing PyPredT6 are
tkinter, time, random, pandas, numpy, csv, sklearn, keras, tensorflow, imblearn,
collections and re.
How to setup PyPredT6
Follow the steps to setup PyPredT6:
- Download the PyPredT6 and its prerequisites from Github:
PyPredT6 Package
. Click the tab "clone or download".
- In the folder you will have the following:
- pypredt6.py - PyPredT6 python script
- samples - folder containing the sample files
- featurefile_eff.csv, featurefile_noneff.csv - the training datasets for effector and non-effector proteins respectively
- Open WinPython-64bit-3.6.1.0Qt5 on Windows and open
the application IDLEX (Python Gui).
- From The GUI click "Open" which will open the file explorer.
- Find the file PyPredT6 (if should be on the same folder as WinPython)
and click "Open" and "Run". On the IDE type "PyPredT6()" and click Enter. A file
entry box will appear.
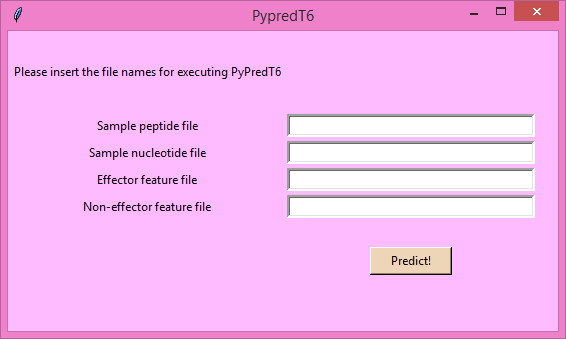
- Find the "Samples" folder from PyPredT6 parent folder.
Under the "Samples" folder, there are three subfolders, "sample1",
"sample2" and "sample3". Each of these folders contain 2 files,
a "gene" file and a "protein" file. The "gene.txt" file contains the sample nucleotide
sequences while the "protein.txt" file contains the sample amino acid sequences in fasta format.
One can create such files for predicting effector proteins among the sequences
provided in these files.
- For field "Sample peptide file" (in fasta format) give the path for amino acid
sequence file of the proteins to be predicted.
- For field "Sample nucleotide file" (in fasta format) give the path for nucleotide
sequence file of the corresponding protein sequence to be predicted.
- For field "Effector feature file" (in CSV format) give the path for effector
feature set file ( featurefile_eff.csv ) for the training.
- For field "Non-effector feature file" (in CSV format) give the path for non-effector
feature set file ( featurefile_noneff.csv ) for the training.
- Fill the fields and click 'Predict'. Wait for the results.
Biological validation of prediction using PyPredT6
File containing the list of the putative effector proteins in Vibrio cholerae and Yersinia pestis
hinting their association with T6 secretion system mediated pathogenesis supported by previously published articles
from the literature can be obtained here. The excel files contain the
Protein names, Uniprot Entry ids, Uniprot Entry names, Gene names,
Gene Ontology (GO) - Biological Process, GO - Cellular Components,
GO-Molecular Function Evidences and their respective DOI.
- Vibrio cholerae
- Yersia pestis

